Since the onset of the coronavirus disease 2019 (COVID-19) pandemic, which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), several new variants have emerged due to multiple genomic mutations. These variants have been classified as variants of concern (VOC) and variants of interest (VOI) by the World Health Organization (WHO).
At present, the SARS-CoV-2 Omicron variant has become the dominant circulating strain in many countries across the world as a result of its high transmissibility as well as its capability to evade immune protection induced by both natural infection and vaccination.
Study: Tracking SARS-CoV-2 Omicron diverse spike gene mutations identifies multiple inter-variant recombination events. Image Credit: CROCOTHERY / Shutterstock.com
The SARS-CoV-2 Omicron variant
The SARS-CoV-2 Omicron variant (B.1.1.529) was initially identified in South Africa in November 2021 and was quickly detected in many countries around the world. Researchers have classified the Omicron variant into three sublineages including BA.1, BA.2, and BA.3.
Over thirty mutations in the spike (S) protein of the Omicron strain have been identified as compared to the original SARS-CoV-2 strain. These mutations have been found to modify the immunogenicity and binding efficiency of the Omicron variant.
Similar to the Omicron variant, previous VOCs including the Alpha (B.1.1.7) and Delta (B.1.617.2) variants also consist of several spike gene mutations that have contributed to the enhanced transmissibility of these strains of SARS-CoV-2.
SARS-CoV-2 recombination
One of the primary concerns regarding the evolution of SARS-CoV-2 is the possibility of developing recombinants, which have often been described as “Demicron” or Deltacron,” that arise from different variants replicating simultaneously in a host. Often, scientists have debated whether a new variant is true, recombinant, or a result of sequencing error.
The process of recombination expedites virus evolution through genes. Recombination could cause changes in phenotypes and alterations in the pathogenicity profiles, host species virulence, zoonotic and anthroponotic transmission, and host adaptation.
Although recombination events associated with other coronaviruses like SARS-CoV-1 and the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) are well documented, it has been difficult to identify recombination signatures in SARS-CoV-2 variants due to its high degree of similarities with SARS-CoV-2 isolates.
About the study
In a new study published on the bioRxiv* preprint server, researchers demonstrate that newly emerging and circulating Omicron subvariants have originated partially through recombination with other variants. To this end, the researchers studied the diversity in the spike protein of the Omicron variants, as well as shared spike mutations between Omicron and other VOCs and VOIs.
Taken together, a total of 52,563 high-quality Omicron spike amino acid sequences were obtained from the GISAID database, which included both BA.1 and BA.2 sequences. The researchers used various computational tools, such as Pymol 2.0, TBTools, BioEdit, BioAider, and jvenn, to analyze the sequences, whereas NextClade was used to develop and annotate whole-genome phylogenetic trees.
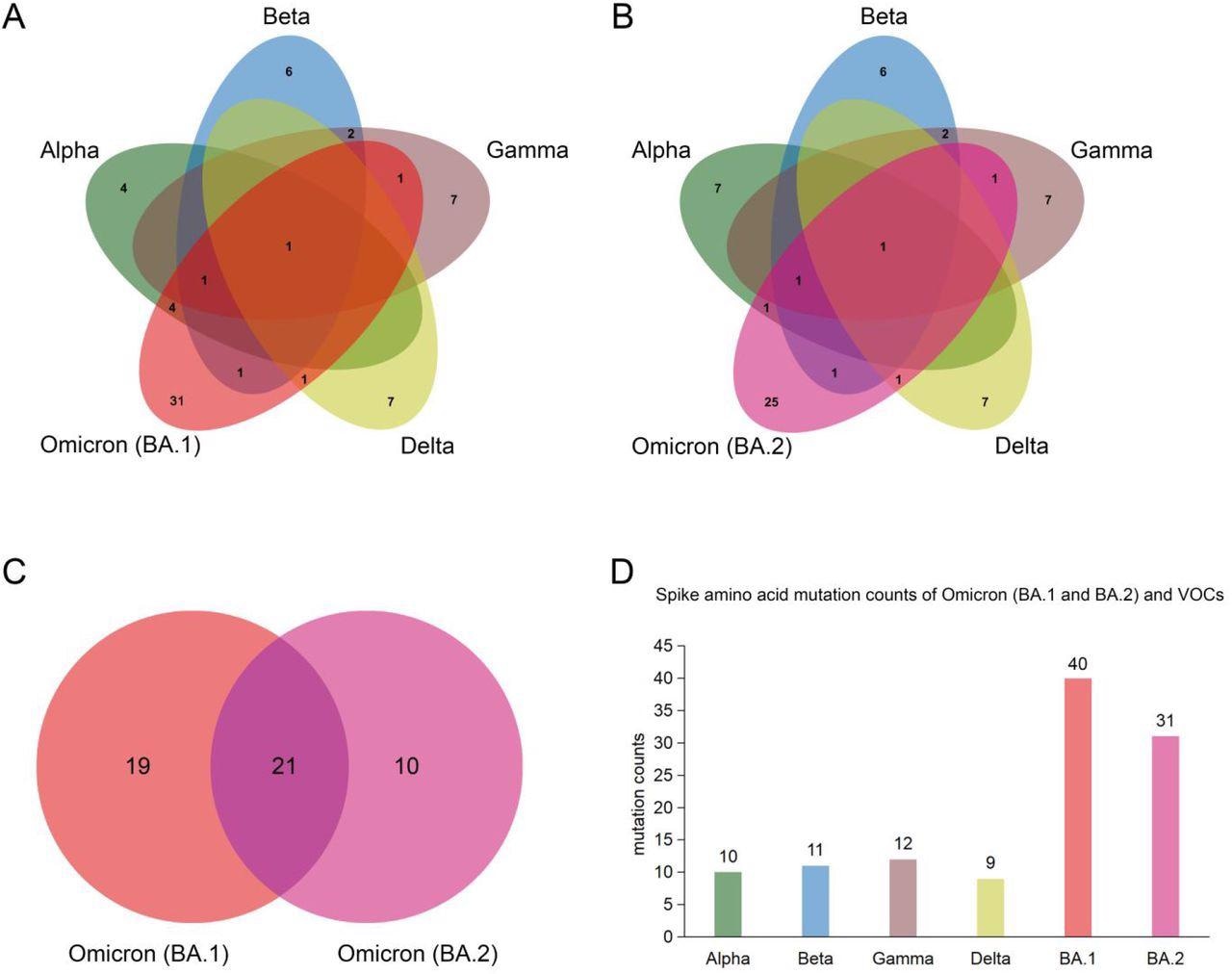
Spike protein amino acid mutations of the Omicron subvariants (BA.1 and BA.2) compared with mutations from the other four variants of concern (VOCs). (A) Venn diagram noting mutations of Omicron (BA.1) and those of VOCs. (B) Venn diagram of Omicron (BA.2) mutations compared to ones of VOCs. (C) Venn diagram of mutations between Omicron (BA.1) and Omicron (BA.2). (D) Spike protein amino acid mutation counts of Omicron (BA.1 and BA.2) subvariants compared with mutations of VOCs.
Study findings
Among the two Omicron variants, BA.2 is rapidly replacing BA.1 and is becoming the dominant epidemic variant in many countries. In this study, the researchers identified common mutations among the BA.1 and BA.2 variants and other SARS-CoV-2 VOCs. Taken together, the BA.1 and BA.2 subvariants were found to share twenty-one spike amino acid mutations that include one in the N-terminal domain (NTD), twelve in the receptor-binding domain (RBD), four in the subdomain (SD).
The BA.1 subvariant shares six, three, three, and two common amino acid mutations with the Alpha, Beta, Gamma, and Delta VOCs, respectively. This sharing of mutations indicates a possible origin of the Omicron variant from these VOCs. The scientists have also identified eight novel mutations in the BA.1 isolate that are present in other VOCs and VOIs. Comparatively, the BA.2 shares only six amino acid mutations with most VOCs.
Upon studying the Omicron subvariants’ spike protein haplotypes, the spike gene of these subvariants was found to consist of forty-nine representative haplotypes. A large number of nucleotide mutations were also identified in the haplotypes compared with BA.1 and BA.2.
The researchers also studied phylogenetic networks to detect possible recombination events among highly similar Omicron variants or subvariants. Several models have revealed that pathogenicity can be increased as a result of virus co-infection and recombination. In fact, one previous study revealed that in a patient coinfected by both the Beta and Delta variants with a 1:9 ratio after fourteen days of infection, recombination in the Orf1ab and spike genes occurred. These events might not be rare, as multiple variants are circulating at a given time.
Conclusions
The authors of the present study revealed that several recombination events occurred between two Omicron subvariants and several VOCs and VOIs. This finding indicates that both co-infection and subsequent genome recombination play an important role in the evolution of SARS-CoV-2. Continual genomic surveillance is essential to determine SARS-CoV-2 mutations and their impact on humans.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Ou, J., Lan, W., Wu, X., et al. (2022). Tracking SARS-CoV-2 Omicron diverse spike gene mutations identifies multiple inter-variant recombination events. bioRxiv. doi:10.1101/2022.03.13.484129. https://www.biorxiv.org/content/10.1101/2022.03.13.484129v1
Posted in: Genomics | Medical Science News | Medical Research News | Disease/Infection News
Tags: Amino Acid, Coronavirus, Coronavirus Disease COVID-19, covid-19, Evolution, Gene, Genes, Genetic Recombination, Genome, Genomic, MERS-CoV, Mutation, Nucleotide, Omicron, Pandemic, Protein, Receptor, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Virus

Written by
Dr. Priyom Bose
Priyom holds a Ph.D. in Plant Biology and Biotechnology from the University of Madras, India. She is an active researcher and an experienced science writer. Priyom has also co-authored several original research articles that have been published in reputed peer-reviewed journals. She is also an avid reader and an amateur photographer.
Source: Read Full Article