In a recent study posted to the bioRxiv* preprint server, researchers analyzed antigenic variety across the significant severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants, like Omicron lineages.
 Study: Post-vaccination Omicron infections induce broader immunity across antigenic space than prototype mRNA COVID-19 booster vaccination or primary infection. Image Credi: Corona Borealis Studio / Shutterstock
Study: Post-vaccination Omicron infections induce broader immunity across antigenic space than prototype mRNA COVID-19 booster vaccination or primary infection. Image Credi: Corona Borealis Studio / Shutterstock
Background
There is an immediate requirement for coronavirus disease 2019 (COVID-19) vaccination techniques to provide the broadest protection against current and future SARS-CoV-2 variants. More than 6.34 million fatalities and 553 million infections have been attributed to SARS-CoV-2 globally.
Even when population immunity persists to elevate because of infections, primary series vaccination, reinfections, or booster vaccination, SARS-CoV-2 is still spreading worldwide. Although SARS-CoV-2 vaccines significantly immunize from severe COVID-19, novel emerging viral variants such as Omicron continue to jeopardize their efficacy even after booster vaccination.
About the study
In the present work, the researchers evaluated the antigenic diversity of SARS-CoV-2 variants and analyzed neutralizing antibody responses to single and numerous exposures across longitudinal infection and vaccination cohorts.
The team employed antigenic mapping to evaluate the antigenic heterogeneity across the major SARS-CoV-2 variants like Omicron BA.2.12.1, BA.4/BA.5, and BA.2 using well-defined sera from a longitudinal group of primary COVID-19 patients with sequence-validated variant infection backgrounds. They used sera from COVID-19-infected subjects in epidemiology, immunology, and clinical characteristics of emerging infectious diseases with pandemic potential (EPICC) research. Indeed, the 1240 primary natural infection neutralizing antibody level readings were analyzed using antigenic cartography to estimate the breadth of immunity among virus variants.
A comparable assessment from an independent group of COVID-19 uninfected people pre- and post-second and third SARS-CoV-2 messenger ribonucleic acid (mRNA) vaccinations supplemented the initial analysis. The scientists quantified the neutralization breadth of sera procured from 39 healthcare professionals in the prospective assessment of the SARS-CoV-2 seroconversion (PASS) research following the receipt of two and three COVID-19 mRNA vaccine doses. They then assessed changes in immunodominance after infection and vaccination using antibody landscapes and other associated methods to deduce antigenic space.
Results
Overall, the researchers found that the antigenic maps of the pre-Omicron SARS-CoV-2 variants were consistent with the antigenic maps of SARS-CoV-2 reported previously. The initial SARS-CoV-2 variants had robust clustering with common amino acid locations. This inference suggests that the particular amino acid alterations regulate the antigenic pattern.
The Omicron BA.2 variant's location on the current map was consistent with an experimental animal antigenic model but not with a map created using antisera from a natural infection. The team also demonstrated that the Omicron BA.4/BA.5 and BA.2.12.1 variants were linked strongly to the Beta cluster, which might shed light on their associated epitopes.
The study findings illustrated that immunodominance hierarchies differ among variants, which might impact the selection of novel vaccine antigens. Likewise, the Delta variant had an identical antigenic distance to the original strain after the third vaccine shot. On the contrary, the Mu and Beta migrated closer to the ancestral viral sequence.
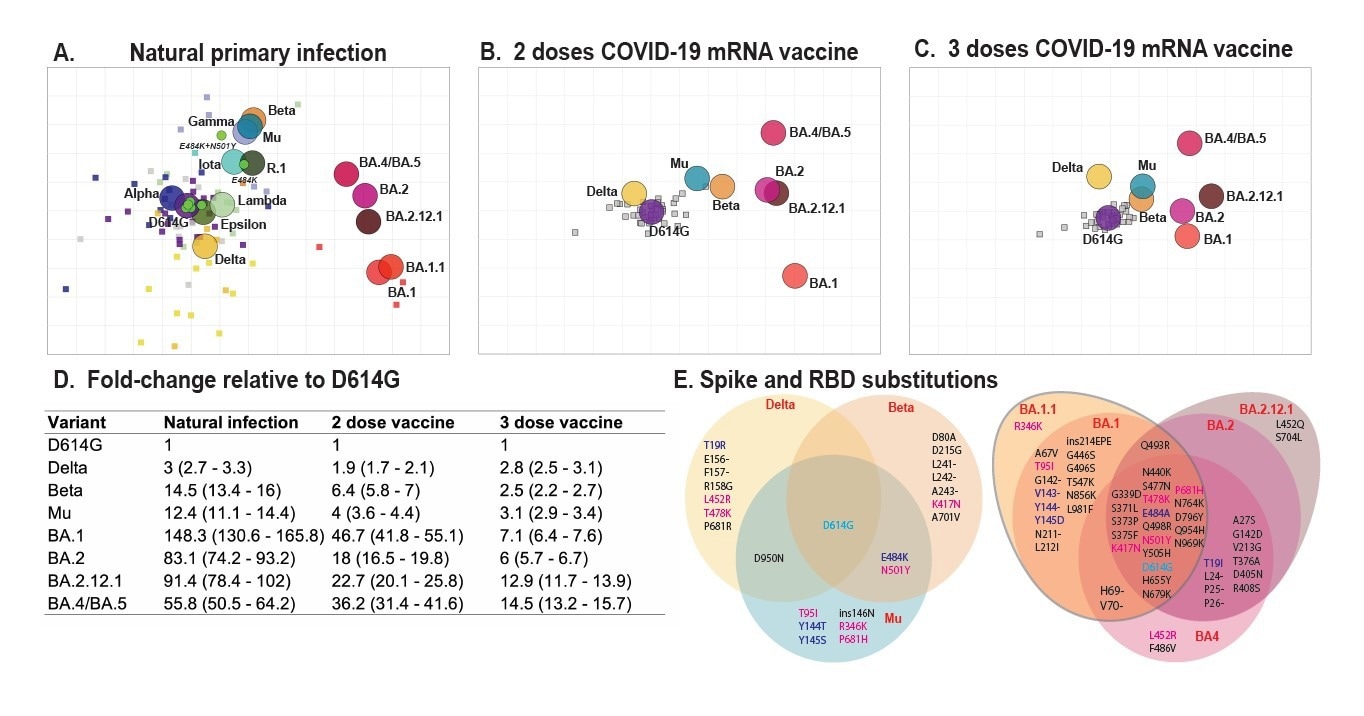
Antigenic maps made with neutralizing antibody titers from single-antigen exposure sera demonstrate BA.1, BA.2, BA.2.12.1, and BA.4/BA.5 are most antigenically distinct from other VOCs. Antigenic maps were made using antigenic cartography with titers for A) sera collected after convalescent primary infection with distinct VOCs and sera from uninfected individuals who received B) 2 doses or C) 3 doses of WT mRNA COVID-19 vaccines. Each grid-square side corresponds to a two-fold dilution in the pseudovirus neutralization assay. Antigenic distance is measured in any direction on the grid. Antigens are shown as circles and are labeled. Sera are shown as squares and are colored by infecting variant. D) Fold-difference in neutralization with 95% confidence intervals from the ancestral strain to each other variant on each map. For example, a fold-difference of four corresponds to two grid squares on the antigenic map. E) Substitutions in the spike and receptor-binding domains for all variants used in this study.
The authors discovered that recurrent exposure to the initial SARS-CoV-2 strain induced by an mRNA vaccine widened immunity to priorly circulating and Omicron variants following the third dose, implying robust back-boosting. In contrast to people who had only received the ancestral variant vaccination, those who received two vaccine doses and then had Omicron post-vaccine infection (PVI) had significantly flatter landscapes, indicating broader protection against both variants.
Additionally, the team noticed that receiving three vaccine doses followed by Omicron PVI induced more robust immune responses than receiving three vaccinations alone. This inference suggests that people who have already got the third vaccine dose centered on the ancestral strain may still be able to expand their immunity by an Omicron-based vaccine.
At least transiently after vaccination, the immunity of people who received a third dose vaccination followed by Omicron PVI was comparable to those who received only two doses and Omicron PVI. Notably, individuals who received two or three doses of vaccine and an Omicron PVI exhibited increased antibody breadth across numerous variants, including Delta. Delta is located in the lower, farther-off region of the antigenic map, indicating that Omicron PVI may have boosted responses to earlier variants.
Additionally, the breadth gain plot analyses revealed that all vaccinated and PVI cohorts had lower responses to BA.2.12.1 and BA.4/BA.5 than predicted based on assessed antigenic distances contrary to the cone-based landscapes ad in line with the antigenic assessments.
Conclusions
The study findings demonstrated how, over time, the antigenic co-evolution of SARS-CoV-2 and its immune reaction in the host human population become increasingly complex. The team offers techniques for characterizing the range of immune reflexes to COVID-19 following various antigenic exposures.
The scientists showed that Omicron PVIs typically result in higher immunity than boosting with vaccine-based on the ancestral strain. Further exposure to the original and Omicron BA.1 antigens can augment antibody coverage against BA.4/BA.5. Although BA.4 was antigenically nearer to the ancestral strain, the breadth acquired against BA.4/BA.5 was lower than BA.1, implying complex immunodominance trends.
Developing vaccination methods against forthcoming SARS-CoV-2 strains requires an in-depth understanding of the mechanisms underlying these immunodominance transitions and a thorough assessment of immune breadth. Furthermore, scientific discoveries into the development of antigenic heterogeneity for SARS-COV-2 might also give insight regarding earlier antigenically complex illnesses by demonstrating how the current immunodominance trends and sophisticated landscapes have originated.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Post-vaccination Omicron infections induce broader immunity across antigenic space than prototype mRNA COVID-19 booster vaccination or primary infection; Wei Wang, Sabrina Lusvarghi, Rahul Subramanian, Epsi Nusrat, Richard Wang, Emilie Goguet, Anthony Fries, Fernando Echegaray, Russell Vassell, Si'Ana Coggins, Stephanie Richard, David Lindholm, Katrin Mende, Evan Ewers, Derek Larson, Rhonda Colombo, Christopher Colombo, Janet Joseph, Julia Rozman, Alfred Smith, Tahaniyat Lalani, Catherine Berjohn, Ryan Maves, Milissa Jones, Rupal Mody, Nikhil Huprikar, Jeffery Livezey, David Saunders, Karine Hollis-Perry, Gregory Wang, Anuradha Ganesan, Mark P. Simons, Christopher Broder, David Tribble, Eric D. Laing, Brian Agan, Timothy H. Burgess, Edward Mitre, Simon Pollett, Leah C. Katzelnick, Carol D. Weiss. bioRxiv preprint 2022, DOI: https://doi.org/10.1101/2022.07.05.498883, https://www.biorxiv.org/content/10.1101/2022.07.05.498883v1
Posted in: Medical Research News | Medical Condition News | Disease/Infection News
Tags: Amino Acid, Antibody, Antigen, Assay, Coronavirus, Coronavirus Disease COVID-19, covid-19, Efficacy, Epidemiology, Evolution, Healthcare, immunity, Immunology, Infectious Diseases, Omicron, Pandemic, Pseudovirus, Receptor, Research, Respiratory, Ribonucleic Acid, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Vaccine, Virus

Written by
Shanet Susan Alex
Shanet Susan Alex, a medical writer, based in Kerala, India, is a Doctor of Pharmacy graduate from Kerala University of Health Sciences. Her academic background is in clinical pharmacy and research, and she is passionate about medical writing. Shanet has published papers in the International Journal of Medical Science and Current Research (IJMSCR), the International Journal of Pharmacy (IJP), and the International Journal of Medical Science and Applied Research (IJMSAR). Apart from work, she enjoys listening to music and watching movies.
Source: Read Full Article