The therapeutic potential of microtubule-associated protein targets for cancer therapy is a largely unexplored research area due to a lack of target-specific agents. Sushmita Chatterjee and a research team in nanomedicine, materials science, nanotechnology and biology at the Tel Aviv University in Israel studied the therapeutic capacity of cytoskeleton-associated protein 5, hitherto abbreviated as CKAP5. The protein is associated with microtubules, and Chatterjee and colleagues silenced the gene via short interfering RNA (siRNA) a molecular biological mechanism to study genes, targeting the CKAP5 encapsulated in lipid nanoparticles for in vivo delivery.
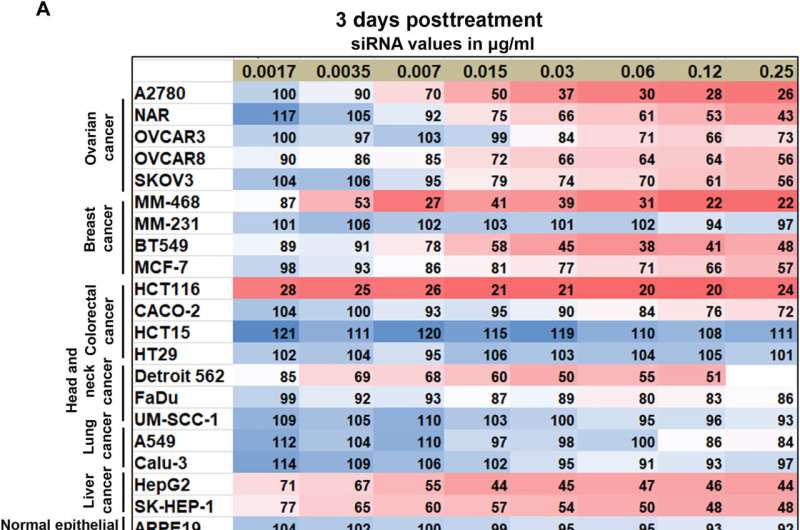
The researchers screened 20 solid cancer cell lines relative to gene silencing to identify a highly responsive chemo-resistant ovarian cancer cell line that underwent significant depletion in mitotic spindle-dynamics for effective experimental cancer treatment. The team showed the therapeutic potential in an ovarian cancer model with an 80% survival rate of silenced-CKAP5 lipid nanoparticle-treated animals. The study is published in Science Advances.
The outcomes highlighted the importance of the gene of interest as a therapeutic target to investigate genetically unstable ovarian cancers to further elucidate its mechanisms of action.
New strategies to treat ovarian cancer
Ovarian cancer is a frequent cause of mortality and accounts for about 4.7% cancer mortality among women. The primary treatment procedure includes maximal surgical resection of the tumor, followed by neoadjuvant chemotherapy with a drug combination. While 60%–80% of patients respond to chemotherapy first, about 80%–85% develop chemo-resistance. As a result, there is a constant search for new therapies to treat ovarian cancer. Scientists have explored several drugs targeting specific receptors although such efforts did not yield effective results for patients with ovarian cancer.
Many chemotherapeutic agents explore defects in the cell cycle machinery of cancer cells to halt the cycle through mitosis inhibition. However, existing mitosis-targeting chemotherapeutic agents do not discriminate between healthy and malignant cell lines, resulting in severe side-effects. The challenge therefore is to identify molecular targets associated with mitosis of cancer cells.
Microtubule-associated proteins (MAPs)
Cell mitosis is an attractive target to effectively treat a variety of cancer forms, where microtubule-associated proteins (MAPs) assist cells to maintain the stability of cell dynamics. One such protein is the cytoskeleton-associated protein 5 (CKAP5), widely expressed in a variety of cells to regulate the dynamics of microtubules in human cells.
In this work, Chatterjee and the team screened the effect of CKAP5 silencing in solid cancer cell lines and in normal non-cancer epithelial cell lines as a negative control. While cells with high genetic instability were selectively susceptible to CKAP5 depletion, a chemo-resistant ovarian cancer cell line showed higher sensitivity to cellular intervention, indicating the gene to be a promising therapeutic target in genetically unstable ovarian cancer cell lines.
High sensitivity to silencing RNA-mediated CKAP5 downregulation in cancer cell lines
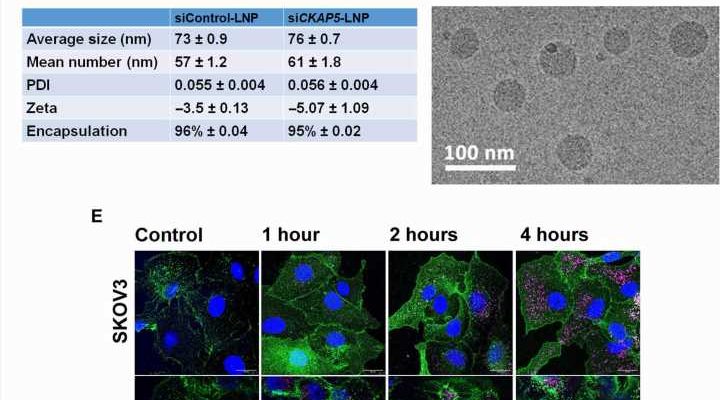
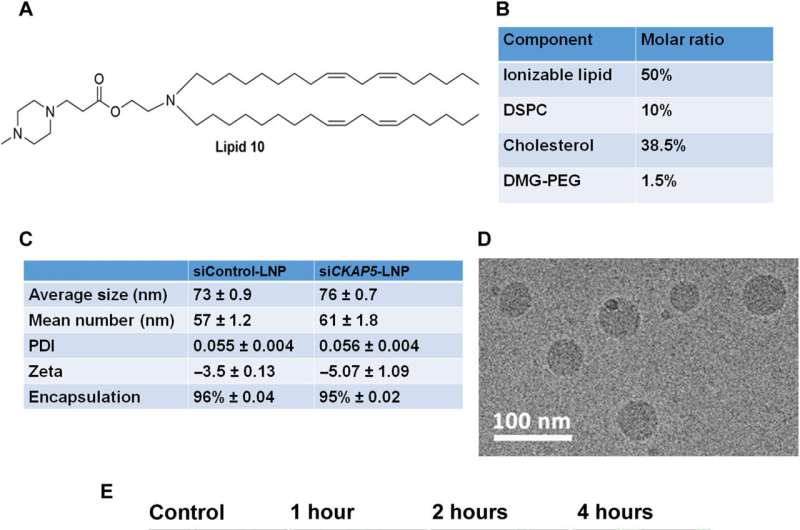
The expression of CKAP5 remains to be well documented in a variety of cancer cells. The researchers first studied the expression of this gene at the transcript level in a panel of solid cancer cell lines, including ovarian, colorectal, lung, and head-neck, while examining its expression in non-cancer cell lines as a negative control. The team used SiRNA-encapsulated lipid nanoparticles, formed via microfluidic mixing, to create uniformly sized structures and examined the outcomes via cryo-electron microscopy.
Of the cell lines tested, they noted eight that were resistant to CKAP5 silencing, including normal or healthy cell lines, which meant that normal cells did not respond toward CKAP5 genetic downregulation. All ovarian cancer cell lines were sensitive to gene silencing; the team therefore exclusively explored this lineage for further studies. While the team downregulated the gene of interest in all cell lines, only specific cancer cells with high genetic instability were vulnerable to CKAP5 depletion.
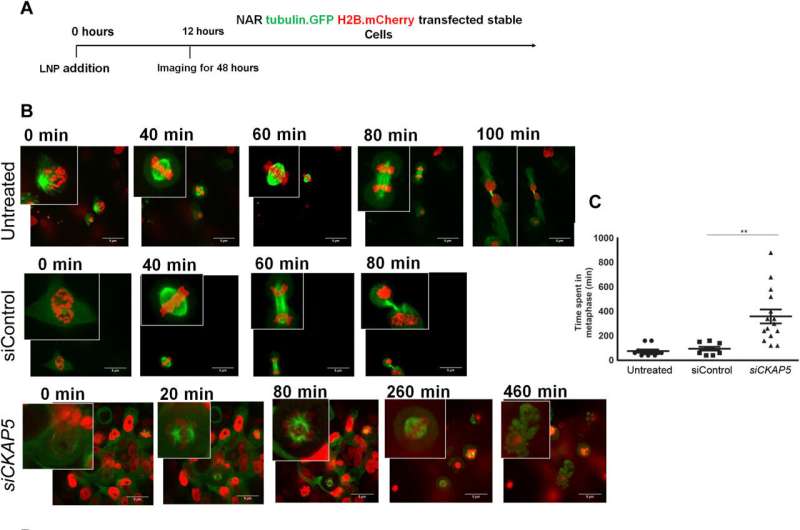
Mechanisms of CKAP5 down-regulation: Cell cycle arrest and spindle defects in ovarian cancer cells
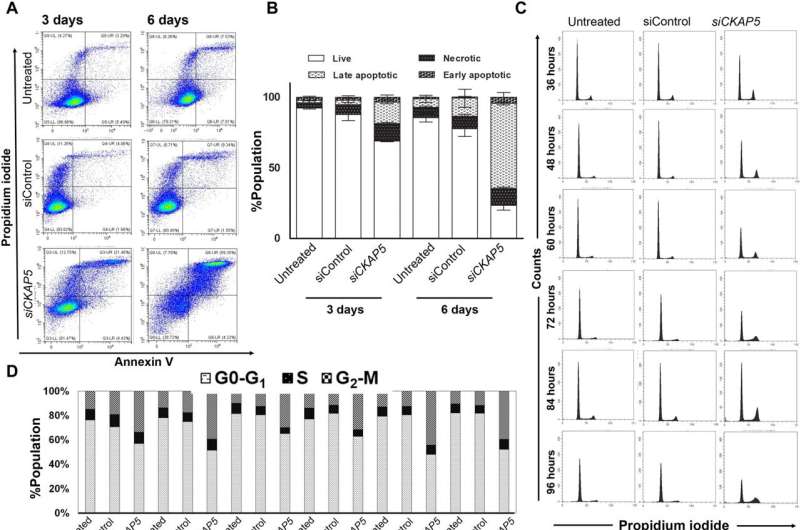
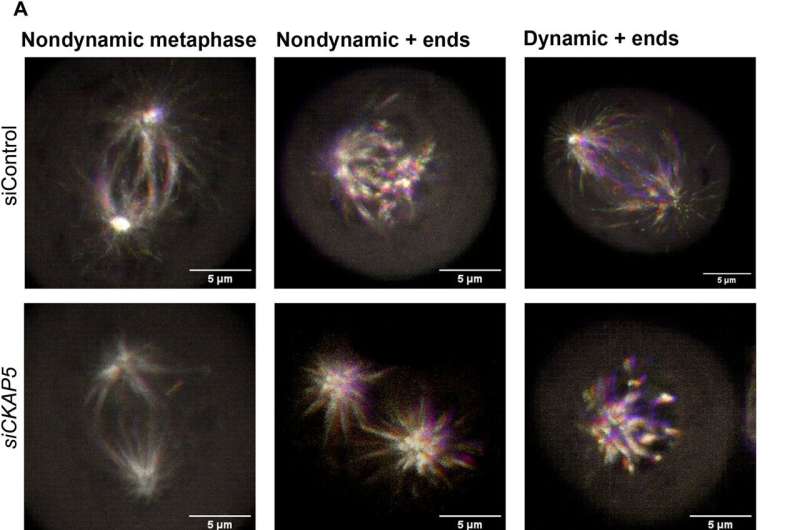
The cancer biologists examined the mechanisms of CKAP5 down-regulation-mediated cell death by developing an apoptotic assay, using propidium iodide-annexin V staining. They noted cell apoptosis (cell death) after transfecting silenced genes into cell lines of interest by using lipid carriers. Spindle damage in response to CKAP5 depletion showed a significant increase in cells arrested in metaphase, compared to control cell lines and untreated cells. The process involved multicentric spindle formation, cell cycle inhibition, and apoptosis, accompanied with spindle checkpoint gene upregulation.
The team studied cell cycle inhibition in two other ovarian cancer cell lines, to show similarities in their behavior to indicate the inhibition of cell viability. When they compared these results in noncancerous normal epithelial cell lines, they remained unaffected in viability and cell cycle state. While all sensitive cancer cell lines underwent increased apoptotic cell death in response to the CKAP5 gene knockdown, there were non-responsive cancer cell lines too that did not undergo apoptotic cell death.
The researchers examined the mechanisms of cell apoptosis in gene-silenced cell lines, to understand the fate of cells. Since the silenced gene was transferred via lipid carriers, they also studied the lipid nanoparticle biodistribution localized in the liver, spleen, and tumor tissues, via fluorescence-activated cell sorting methods.
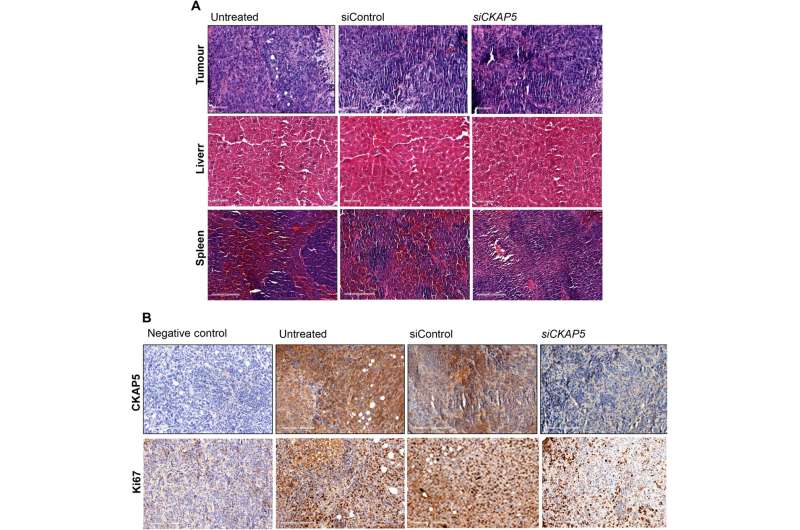
Xenografted tumor growth and increased survival in gene-knockout animal models
The team tested the effects of therapeutic gene silencing in animal models after implanting the genetically modified cell lines into mice to monitor tumor growth. After eight days of implantation, they randomly grouped the mice to treated, control and untreated groups. After administering several doses, they studied the dynamics of tumor growth via fluorescence and luminescence imaging in vivo. After sacrificing the animal models after treatment, they collected the tumor tissues to test in vivo silencing efficiency, and assessed the tissue anatomy. As expected, the animals delivered with the silenced gene yielded substantially reduced genetic expression, when compared to control groups to suggest experimental success.
Outlook
For decades, scientists have targeted the cell cycle to explore cancer therapies to overcome the chromosomal abnormalities and genetic defects associated with cancer cells. At present, tubulin targeting agents are at the forefront of antimitotic agents. However, researchers urgently seek to explore new mitotic targeting agents. In this way, Sushmita Chatterjee and colleagues showed the cytoskeleton-associated protein 5 (CKAP5) gene to play an important role in mitotic spindle assembly by affecting tubulin function. Its inhibition can play a key role to treat genetically unstable ovarian cancer cells.
The study outcomes highlight the vulnerability of cancer cells to CKAP5. Since ovarian cancer is diagnosed at late stages, it will be productive to study the possibility of silencing CKAP5 in late-stage ovarian cancer metastasis, with implications to treat other cancers that metastasize and localize to the intraperitoneal cavity, including pancreatic, liver, and colorectal cancers.
More information:
Sushmita Chatterjee et al, Therapeutic gene silencing of CKAP5 leads to lethality in genetically unstable cancer cells, Science Advances (2023). DOI: 10.1126/sciadv.ade4800
Deborah K. Armstrong et al, Intraperitoneal Cisplatin and Paclitaxel in Ovarian Cancer, New England Journal of Medicine (2006). DOI: 10.1056/NEJMoa052985
Journal information:
New England Journal of Medicine
,
Science Advances
Source: Read Full Article